# Title
Deep-learning-based gene perturbation effect prediction does not yet outperform simple linear baselines
# 数据集
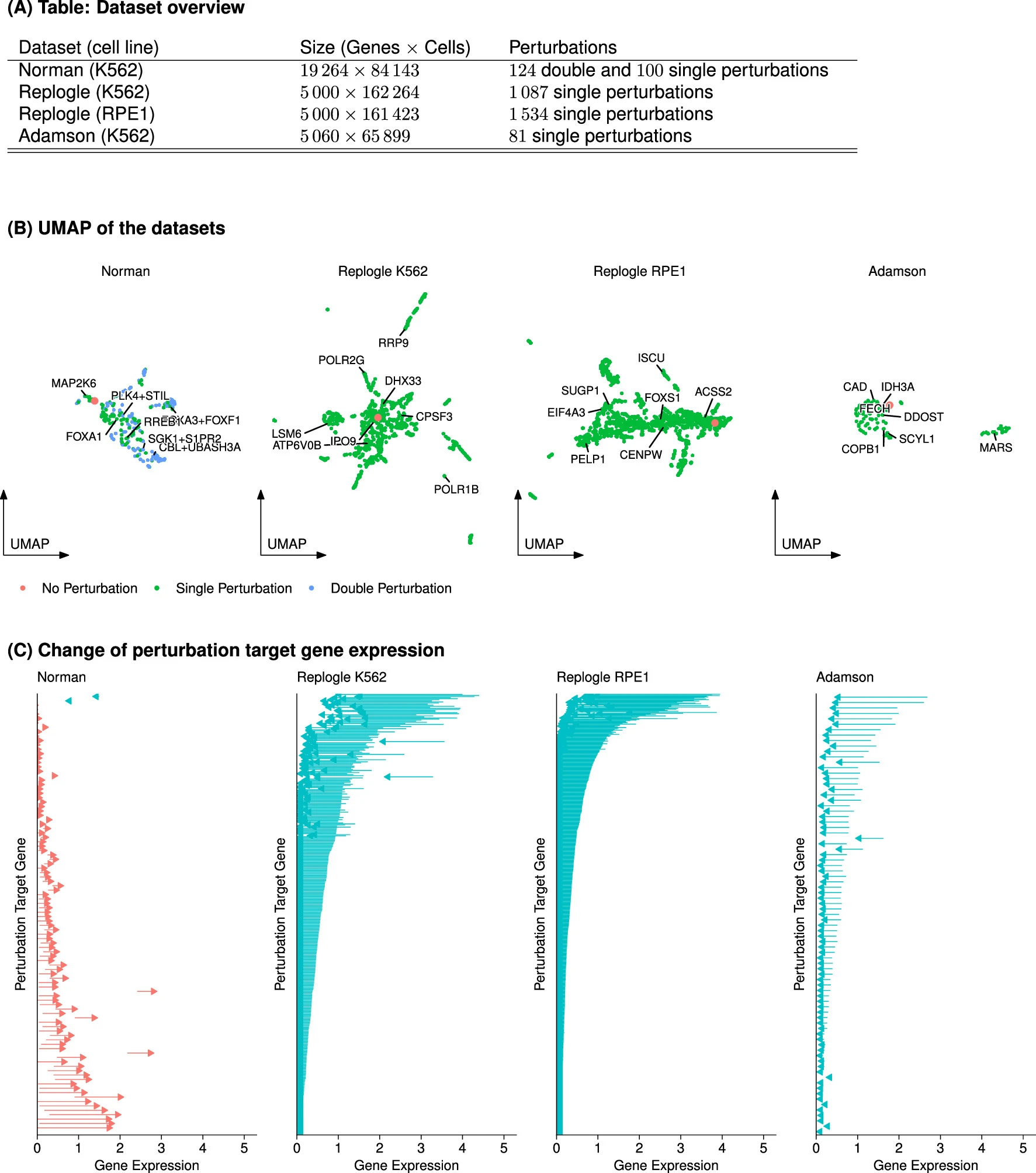
Norman et al.11, in which 100 individual genes and 124 pairs of genes were upregulated in K562 cells with a CRISPR activation system
we used two CRISPR interference datasets by Replogle et al.13 obtained with K562 and RPE1 cells and a dataset by Adamson et al.14 obtained with K562 cells
# 实验表现
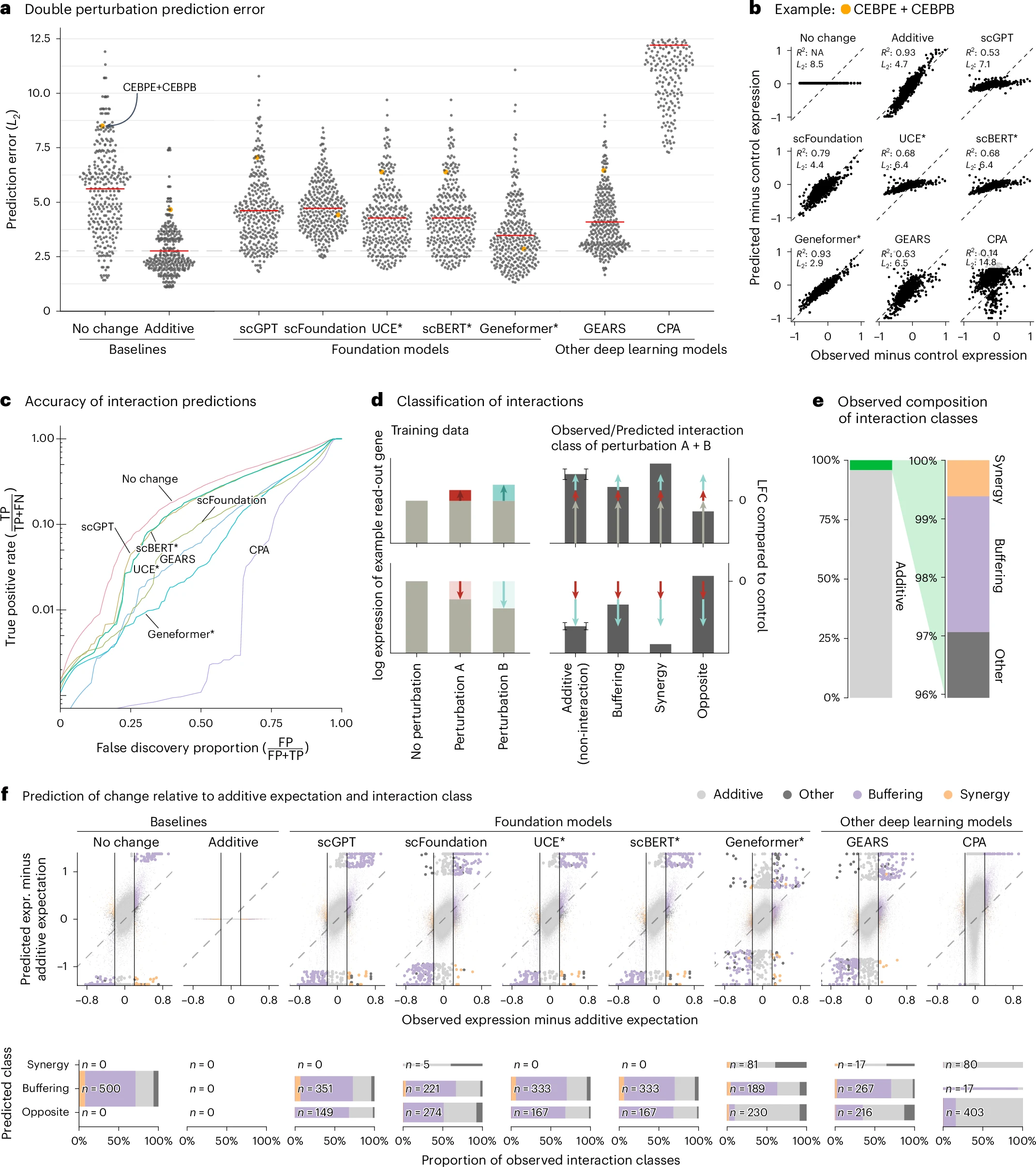
# All models had a prediction error substantially higher than the additive baseline
# Distribution of the observed difference from the additive model
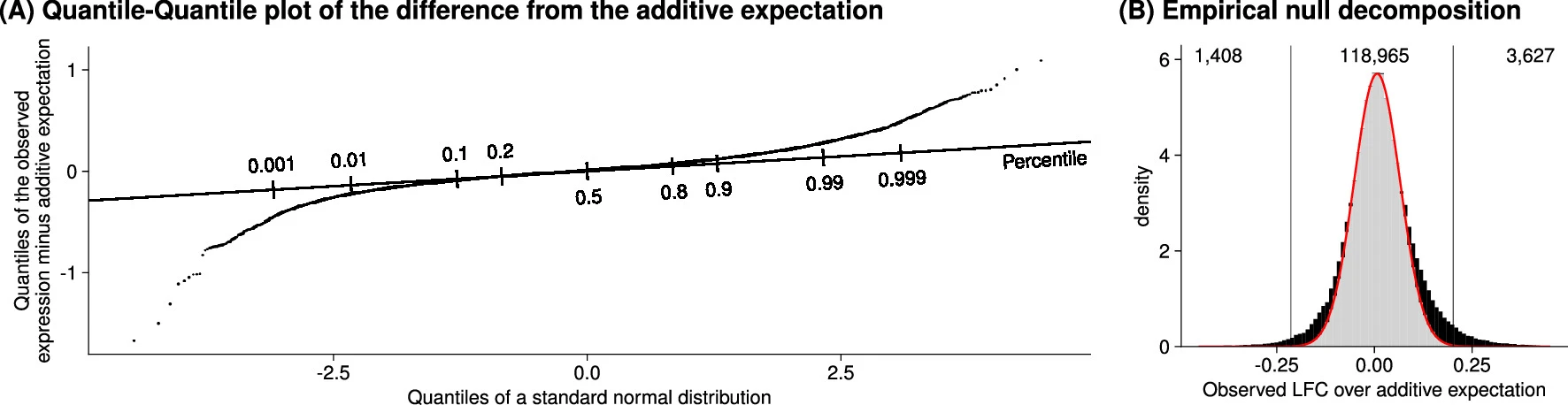
展现了观测值减去零假设期望值的差异分布。这里的零假设模型是仅考虑加性效应的基线模型。
we identified 5,035 genetic interactions (out of potentially 124,000) at a false discovery rate of 5%.
# Variation of the predicted and observed expression values.
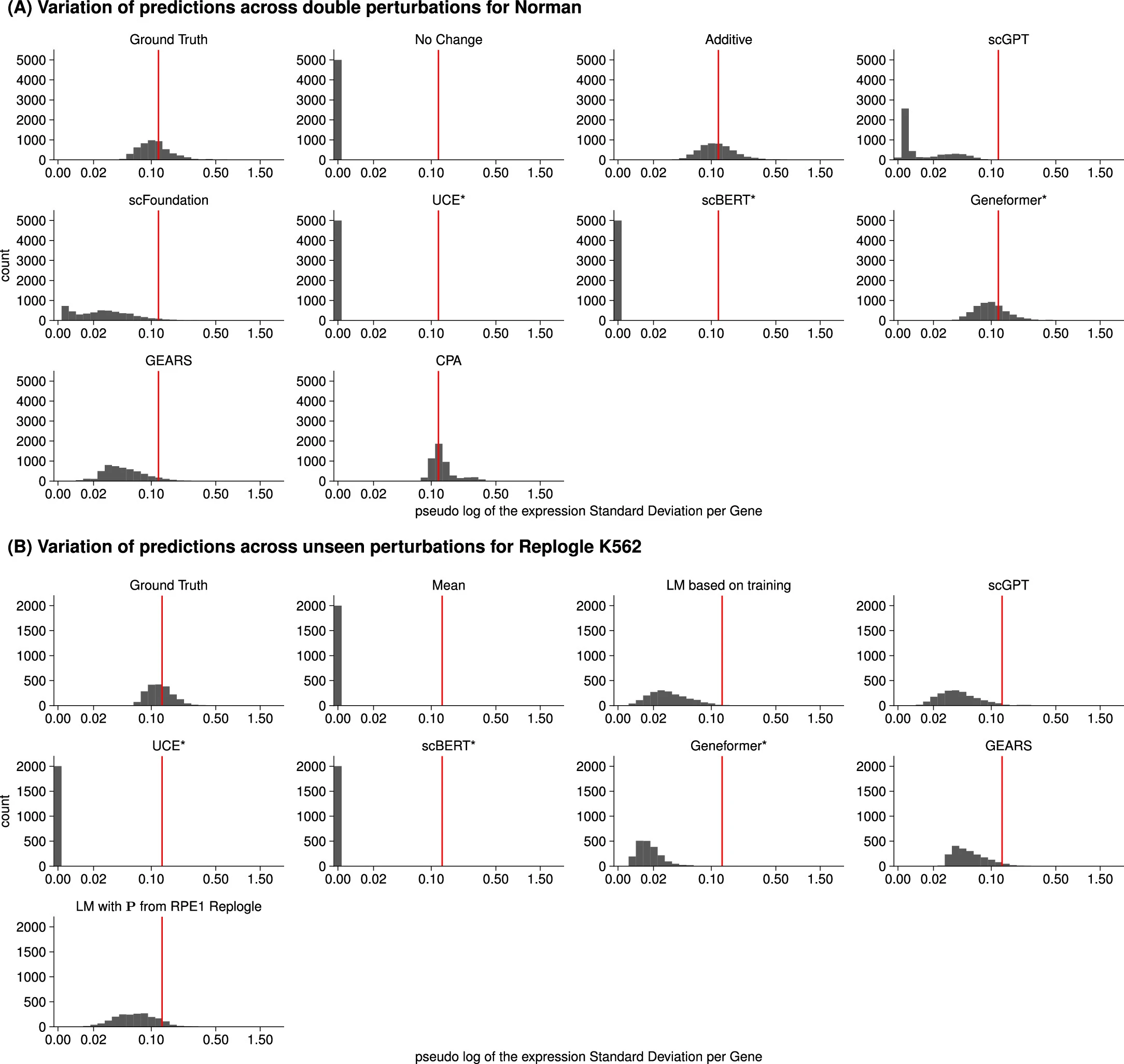
the predictions of scGPT, UCE and scBERT did not vary across perturbations, and those of GEARS and scFoundation varied considerably less than the ground truth
# 设计的一个简单的线性模型作为 baseline
$$\begin{equation} \mathop{{\rm{argmin}}}\limits_{{\bf{W}}}| | {{\bf{Y}}}_{{\rm{train}}}-({\bf{G}}{\bf{W}}{{\bf{P}}}^{T}+{\boldsymbol{b}})| {| }_{2}^{2} \end{equation}$$是 的行的平均值。
# 实验效果
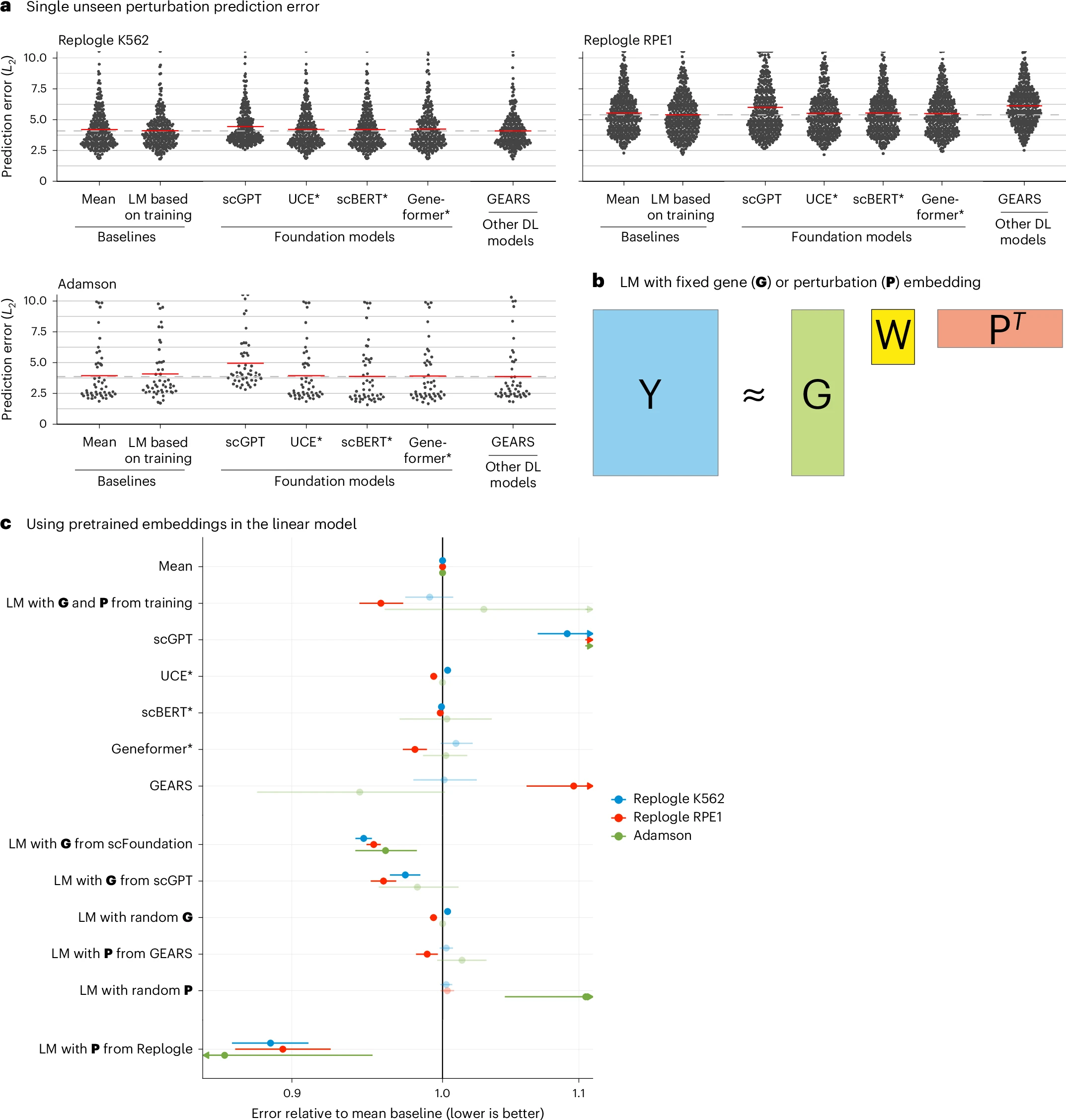
None of the deep learning models was able to consistently outperform the mean prediction or the linear model
# 计算 的值
利用一个带正则化的双边回归公式:
$$\begin{equation} {\bf{W}}={({{\bf{G}}}^{T}{\bf{G}}+\lambda {\bf{I}})}^{-1}{{\bf{G}}}^{T}({{\bf{Y}}}_{{\rm{train}}}-{\boldsymbol{b}}){\bf{P}}{({{\bf{P}}}^{T}{\bf{P}}+\lambda {\bf{I}})}^{-1} \end{equation}$$