这文章看的人心力憔悴
# 单词
这里面的专有名词太多了,会记录很多
单词表
phenotypic: 表型的
mammalian: 哺乳动物的
unicellular: 单细胞的
multicellular: 多细胞的
reproducible: 可再现的
unprecedented: 前所未有的
transcriptomics: 转录组学
granularity: 粒度
pertaining: 有关
modality: 方式
morphological: 形态学的
physiological: 生理学的
heterogeneous: 各种各样的
concordance: 一致的
hurdle: 障碍
untangle: 解开
completeness: 完整
cortical: 外皮的
in vivo: 体内
electrode: 电极
anatomical: 解剖学的
epigenomics: 表观基因组学
delineate: 刻画
feasible: 可行的
Drosophila: 果蝇
C.elegans: 一种线虫
primates: 灵长动物
cortex: 脑皮层
hippocampus: 海马体
striatum: 脑纹体
thalamus: 丘脑
hypothalamus: 下丘脑
cerebellum: 小脑
spinal cord: 脊髓
retina: 视网膜
methylation: 甲基化
situ: 原位
spiced: 粘接
variant: 变体
proteomics: 蛋白质组学
subcellular: 亚细胞的
lock-step: 同步变化
colorimetric: 色度计
connectomics: 连接组学
barcode: 条形码
definitive: 最终的
integrate: 集成的
patched: 写不
retrograde: 逆行
myltiplexed: 多路传输的
Rossetta stone: “罗塞塔石碑”,用来安于要解决一个谜题或困难事物的关键线索
recombinant: 重组的
viral: 病毒性的
coarse: 粗糙的
dysfunction: 机能障碍
iterative: 迭代的
overarching: 总体而言
implementable: 可执行的
phylogenetic: 种系发生的
niches: 生态位
convergent: 相交的
skew: 歪曲
putative: 推定的
bona fide: 善意的
surmount: 克服
annotated: 注释的
homologies: 相似
manifest: 显现
regional: 地区
phylogenomics: 基因组系统学
arguably: 可论证的
hierarchical: 分等级的,等级制度的
comprise: 组成
telencephalon: 大脑
diencephalon: 间脑
mesencephalon: 中脑
rhombencephalon: 后脑
isocortex: 等皮质
olfactory: 嗅觉
cerebral nuclei: 脑核
cortical subplate: 皮质亚板
forebrain: 前脑
tectum: 头盖
tegmentum: 被盖
pons: 脑桥
medulla: 髓质
associational: 联想的
neurotransmitters: 神经递质
glutamatergic: 谷氨酸能
GABAergic: 氨基丁酸能
axon: 轴突
inhibitory: 抑制的
canonical: 典型的
dichotomy: 一分为二
subcortical: 皮层下
ganglionic: 神经节的
eminence: 突出
elaborate: 复杂的
coexistence: 和平共处
spiny: 多刺的
projection: 投射
vascular: 血管的
endothelial: 内皮细胞的
neuroectoderm: 神经外胚层
astrocytes: 星形胶质细胞
glia: 胶质细胞
oligodendrocytes: 少突胶质细胞
trajectory: 轨迹
heterogeneity: 异质的
ambiguity: 模棱两可的
laminar specificity: 层特异性
cis-regulatory: 顺式调控
assay: 检验
coupling: 联系
somatosensory: 体感
responsive: 反应灵敏的
pathological: 病理学的
metabolic: 新陈代谢的
pharmacological: 药物学的
lineage: 血统
maturation: 成熟
progenitors: 祖先
coordinate: 协调
perceptual: 感知的
screen: 筛查
immunostaining: 免疫染色法
transgenic: 转基因的
ion channels: 离子槽
induce: 诱发
vulnerability: 易遭攻击
perturbation: 不安です
mechanistic: 机械论的
restorative: 促进康复的
baseline: 起点
undergo: 经历
homeostasis: 恒常性
buffering: 缓冲
secretion: 分泌
reactive: 易起化学反应的
plasticity: 可塑性
sclerosis: 僵化
schizophrenia: 精神分裂
cerebrovascular: 脑血管的
integrity: 完整性
innate: 天生的
peripheral: 外围设备的
mesodermal: 中胚层的
yolk sac: 卵黄素
plethora: 过多
relatively: 相对的
neurodegenerative: 神经退化
neuroinflammatory: 神经炎
fertilize: 施肥
zygote: 受精卵
unravel: 解开
cascade: 传递、倾泻
proliferation and differentiation: 增殖和分化
manifestation: 表现
ontogeny: 个体发育
multipotent: 有多种能力的
give rise to: 引起
roll out: 推出
nich: 生态位
dendrite: 树突
circadian: 昼夜节律
synaptogenesis: 突触发生
gliogenesis: 胶质生成
myelination: 髓鞘生成
arborization: 树木化
dorsal: 背部的
ventral: 腹部的
complementary: 互补的
in turn: 依次
such that: 以至
emanate: 发源
adhesion: 粘连
assembly: 聚集
sculpt: 使... 成型
granular: 粗糙的
fuzzier: 模糊的
account for: 占用、解释
rationale: 基本原理
tantalizing: 使... 着迷
nomenclature: 命名系统
# 观点摘抄
A cell is the basic unit of all living organisms(except for viruses)
cells within an organism can be grouped into types. ceks within a type exhibit similar structure and function that are distinct from cells in other type.
It is often unclear if cell types defined by different phenotypic features agree with each other nor which feature is the right one to define cell types.
通常不清楚由不同表型特征定义的细胞类型是否相互一致,也不清楚哪个特征是定义细胞类型的正确特征
It(single-cell transcriptomics) has been used to define cell types in a variety of species, tissue organs, and brain regions.
In many cases, lacking a way to reproducibly label a cell type(typically using a molecular genetic approach)presents a major hurdle to relate different studies and findings to each other.
Currently, single-cell transcriptiomics and connectomics are two primary approaches that have the potential to meet the completeness requirement.
Transcriptomics by single-cell or single-nucleus RNA-sequencing(scRNA-seq or snRNA-seq)is now the most widely used approach to generate cell type taxonomies or atlases from many species, tissue organs, and brain regions, due to its comprehensiveness and high dimensionality
Single-cell epigenomics, such as single-nucleus ATAC-seq(to characterize chromatin accessibility) or DNA methylation-sequencing has also been used to generate cell type atlases for different brain regions that are consistent with transcriptomic cell atlases and further reveal cell type-specific gene and chromatin regulatory landscapes.
Spatially resolved transcriptomics, including a variety of techniques based on in situ imaging, in situ capture, or in situ sequencing, is a powerful approach combing molecular and spatial characterization ar single-cell or near-single-cell level, revealing spatial relationships between cell types both local environment and global architecture.
A cell's morphology and connectivity has been regarded as the most defining feature of brain cell types since the era of Cajal, although its place may be overtaken by transcriptome.
Cell type classification has been compared with species classification.
evolutionary approach based on comparative genomics has brought an entirely new paradigm to species classification, providing a sysmetic, rational, unbiased, universally applicable, and extensible classification scheme.
It has been proposed that the foemation of new cell type identity requires the evolution of a unique cell type regulatory signature that includes a cell type-specific core regulatory complex(CoRC) of transcription factors, which defines the identity and coordinated gene expression pattern of the new cell type.
Many gaps also exist dur to the extinction of intermediate species.
It is possible to gain a deep understanding of cell types from even a single species, since each species has evolved from its simpler ancestors through mant rounds of cellular and regional duplications in which the newly created cell types and regions adopt new functions, and thus, comparing between cell types and between regions within the species can reveal the evolutionary relationships between cell types as well.
Therefore, the first and foremost principle is that cell types are the product of evolution and cell type identities are encoded in the genome.
Evolution of individual cell types maybe more complicated than organismal evolution as a whole, and it will be interesting to see if different cell types evolve in similar or different ways as the whole organism.
A transcriptome also contains gene exppression profiles that underlie arguably all phenotypic features of the cell at the time of state when the cell is characterized.
Transcriptomically derived cell type taxonomies in the adult mammalian brain, with majority of the studies conducted in mouse, have consistently revealed a hierarchical organization of cell types.
The highest level of branches is the separation of neuronal and various non-neuronal cell classes.
There may be cell types crossing or shared between brain structures due to cell migration duuring development.
Transcriptomic cell type taxonomies from visual cortex and motor cortex display a similar organization.
The glutamatergic excitatory neuurons mostly have long-range axon projections to other cortical and/or subcortical regions.
The GABAergic inhibitory neurons mostly have their axon projections confined within the local area.
This orgnization is highly consistent with the existing knowledge about cordical cell types that have been extensively studied ina variety of phenotypic modalities over the past 50 years,suggesting that single-cell transcriptomics alone can faithfully capture the overall cell type orgnization at class and subclass levels,although the validity of transcriptomic clusters at the lowest branch level has yet to be fully tested
We may hypothesize that the adult-stage transcriptomic landscape can reveal the organization of cell types within and between brain regions that reflect their evolutionary and developental histories,
Another prominent feature of the relationship between cell types revealed by transcriptomic studies is the coexistence of discrete and continuous variations between types.
Discrete variations exist among cell sub-classes and major types that are usually ar the higher branches of the hierarchy. Continuous variations are usually found among closely related transcriptomic clusters or subtypes at lower branches,
In brain transcriptomic cell type taxonomies non-neuronal cells generally display less diversity than neurons, with little regional specificity except for astrocytes.
Immature and mature oligodenfrocytes form a long continuous trajectory originating from OPCs, indicating coexistence of multiple states of multiple states of gradually maturing oligodendrocytes
These studies further suggest that the hierarchical organization of brain cell types is a frame-work extensible to describling cell type evolution.
For the categorization of cell types based on their transcriptomes tobe meaningful and to understand their relevance to the structure and function of the tissue organ where the cells reside, it is necessary to chracterize other modalities of celluar properties.
Since a neuron type usually has multiple projection targets and a neuron within that type can choose a subset of those targets either specifically or randomly, a single-target-site Retro-seqe assay is often insufficient to resolve the target specificity of a transcriptomic type except in special cases.Brain-wide complete reconstruction of single-neuron morphology is currently the only approach to capture the full extent of a neuron;s axon projection pattern and define projection neuron types.
extensive heterogeneity among individual neurons; this heterogeneity reflucts three axes of variations: regional specificity, topographic specificity, and individual(potentially stochastic) variation,which do not readily correlate with transcriptomic types within the subclass.
It has been suggested that neuron types may be defined by their unique communication properties implemented as synaptic input-output patterns.
A greater opportunity lies in the Drosophila field where comprehensive catalogs of both transcriptomic cell types and connectional cell types have been obtained independently, and a systematic comparision and cross-correlation between them may be realized soon.
To compare functional properties among transcriptomic cell types, two general approaches have been taken - coupling in vivo calcium imaging with post hoc multiplexed FISH to decode the molecular identities of the imaged cells or mapping immediate early gene(IEG) activation during sensory response or behavior using scRNA-seq or MERFISH.
For a definition of cell types to be meaningful, it must be associated with what cell types do.
Since transcriptomes are rich in containing the molecular correlates of all sorts of cellular properties, specific molecular signatures responsible for certain essential anatomical or func- tional features may be hidden below noise and will need to be brought out through supervised approaches and used to refine the classification of cell types toward more functional relevance. It is also necessary to trace back into development to identify potentially clearer molecular correlates as different connectional or functional properties may be established at different developmental time points. On the other hand, it is also reasonable to propose that some connectional or functional properties should not be used to define cell types because they may be emerging properties arising from the interaction of a network of cell types or from experience and/or activity-dependent processes that represent a cell state rather than a defining feature for a cell type.
A key question arising when evaluating a transcriptomic taxonomy is whether some clusters actually represent a particular cell state a transient or dynamically responsive property of a cell to a context—rather than a cell type, as a cell type can exist in different states.
Although not absolute, it is reasonable to assume that transcriptomic changes tend to be more continuous during cell state transitions and more abrupt or discrete when cells switch their types.
More often, emergence of a new cell type during development is the consequence of cell division from which a daughter cell takes up a new cell type identity
Pharmacological or genetic (e.g., CRISPR-based) perturbations in normal or diseased conditions, in combination with single-cell profiling (e.g., Perturb-seq), are powerful approaches to gain a mechanistic understanding of how disruptive or restorative cell state changes can affect cell type function or dysfunction
Here, I highlight a prominent feature of the non-neuronal cell types in the brain, which is that despite having lower diversity than neurons in baseline adult state, many non-neuronal cell types undergo pronounced (显著的) changes, i.e., they exhibit many different cell states, under different physiological or diseased situations.
Taken together, these studies paint a collective picture on how the variety of non-neuronal cell types actively respond to and contribute to different physiological and pathological changes in the brain.
A deep understanding of a subject often comes from understanding how it is built.
In the developmental ontogeny of cell types, earlier-stage ancestral cell types are fewer and are more multipotent, and they give rise to a larger number of descendant cell types with increasingly restricted fates.
Systematic single-cell transcriptomic, epigenomic and spatially resolved transcriptomic profiling with high temporal resolution, coupled with lineage tracing and other phenotypic characterization, holds tremendous potential to capture key sets of genes and genomic regulatory networks involved in these series of events and begin to resolve the extremely complex spatial and temporal transitions of cell types and states leading to the adult-stage repertoire of cell types
it has been shown in multiple systems that a progenitor can produce cells belonging to several neuronal and non-neuronal types, ordered by developmental timing
In conclusion, cell types are the product of evolution, and they are the basic functional units of an organism.
A hierarchical presentation of cell types is more meaningful than ascertaining an exact number of types.
# 内容概括
# Introduction
概述细胞在不同物种之间存在相似性与不同,细胞分类的困难,新型的 single-cell transcriptomics 带来了全新的视角但仍然在区分细胞的方式上存在问题
# Approaches to characterize cell types
区分细胞的方式很困难,去找到合适的方法去综合的、无偏差的、有质量的、有标准的、可扩展的衡量细胞种类很有必要,接下来介绍了几种方法:
最开始的:physiological and functional 两种方式,存在许多的问题
后来: Molecular and anatomical 方式更好的解决了问题
现在:single-cell transcriptiomics and connectomics 是最主要的两种方式
先介绍了 transcripomics 这方面有多牛,它解决了许多问题,然后介绍了 epigenomics\ Spatially resolved transcriptomics\proteomics 这些在 transcripomics 衍生出来的东西能做到许多事情。介绍了与之不同的 morphology and connectivity 在历史上挺牛,重要性也挺高,但目前在被逐渐取代。最后总结需要用一种集成的方式去判断细胞类型
技术上来说,这里有三种方式去达成这种集成:
- conduct multimodal characterization from the same cell using approaches such as single-cell multi-omics.
- perform label transfer between independently collected datasets through "Rosetta stone" features.
- create type-targeting genetic tools using marker genes, promoters, and enhancer elements identified from transcriptomic and epigenomic cell atlases and use these tools for structual and functional studies.
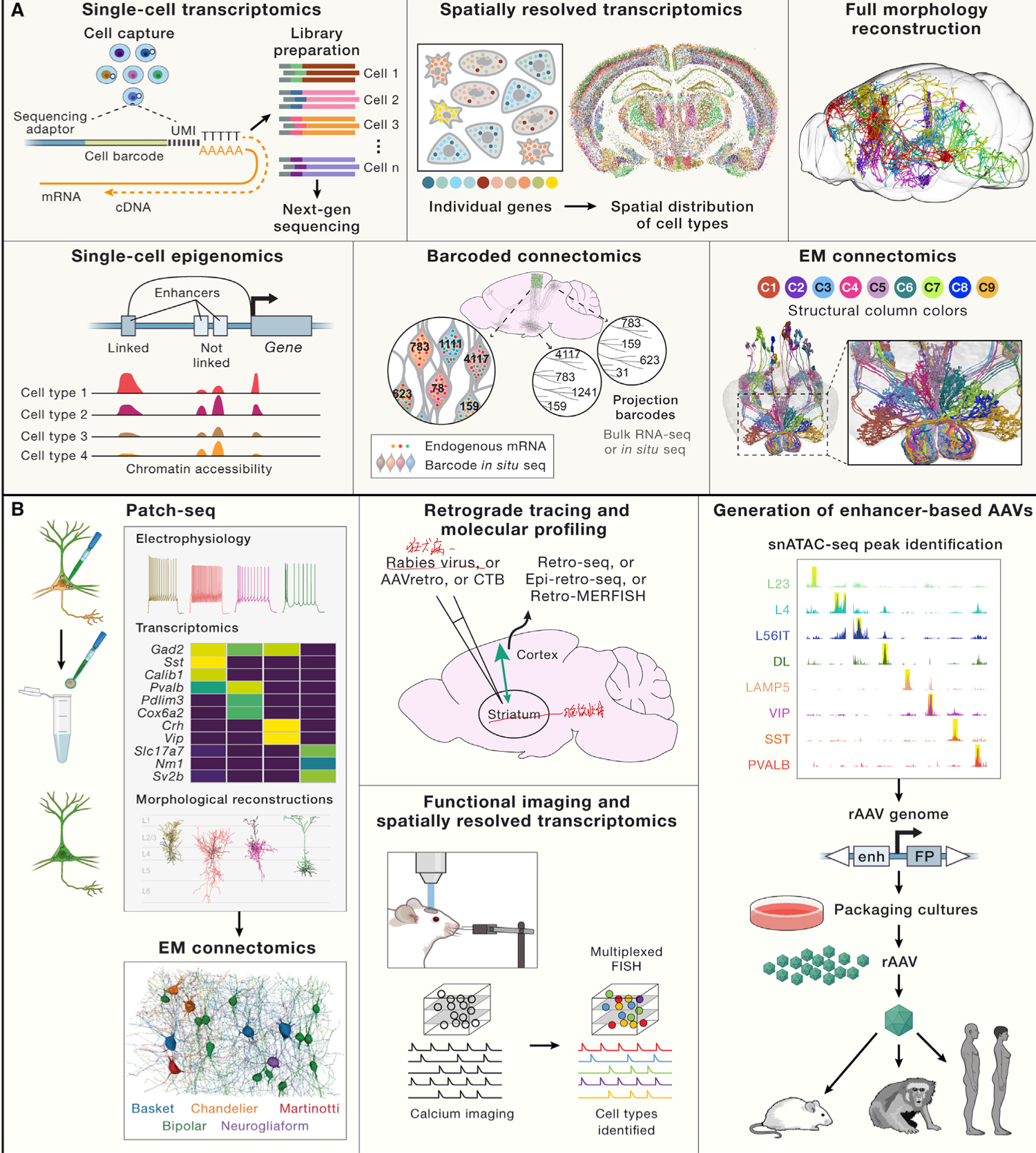
# Cell types are the product of evolution
介绍了物种与细胞分类的相似性,介绍了基于比较基因的 evolutionary approach 的一些方法,这些方法存在哪些困难,并说明了这种方法的好处。
提出了一个观点,细胞的种类由基因决定,具体的是由 CoRC 决定,并给出了证据。
说明达到 evolutionary defination 存在困难,然而我们仍然可以仅仅根据某个 species 去定义细胞类型。
最后论述了 transcriptomes 能够达到的效果。
# Hierarchical organization of transcriptomically defined cell types
首先总引利用 transcriptomic 揭示了细胞类型的分等级的组织架构。
利用哺乳动物的大脑结构做了说明,总体介绍了大脑的结构。
接着利用 cortex 做介绍,分别对比了 glumatergic 和 GABAergic 的性质不同,在不同领域分类方式不同(例如 glumatergic 根据区域不同分为 motor 类和 visual 类,而 GABAergic 在不同区域分类一样),然后根据这一性质做了解释,glumatergic 是由于 gradient expression of morphogens 导致的,而 GABAergic 是由于迁移导致的。然而,在 glumatergic 中,即便分类不同,但它们仍然是 one-to-one homology 的。
接着说明了细胞类型分类中的连续性和离散型的话题。可分为大类,然而个体之间亦存在差别。然后举了一些例子证明。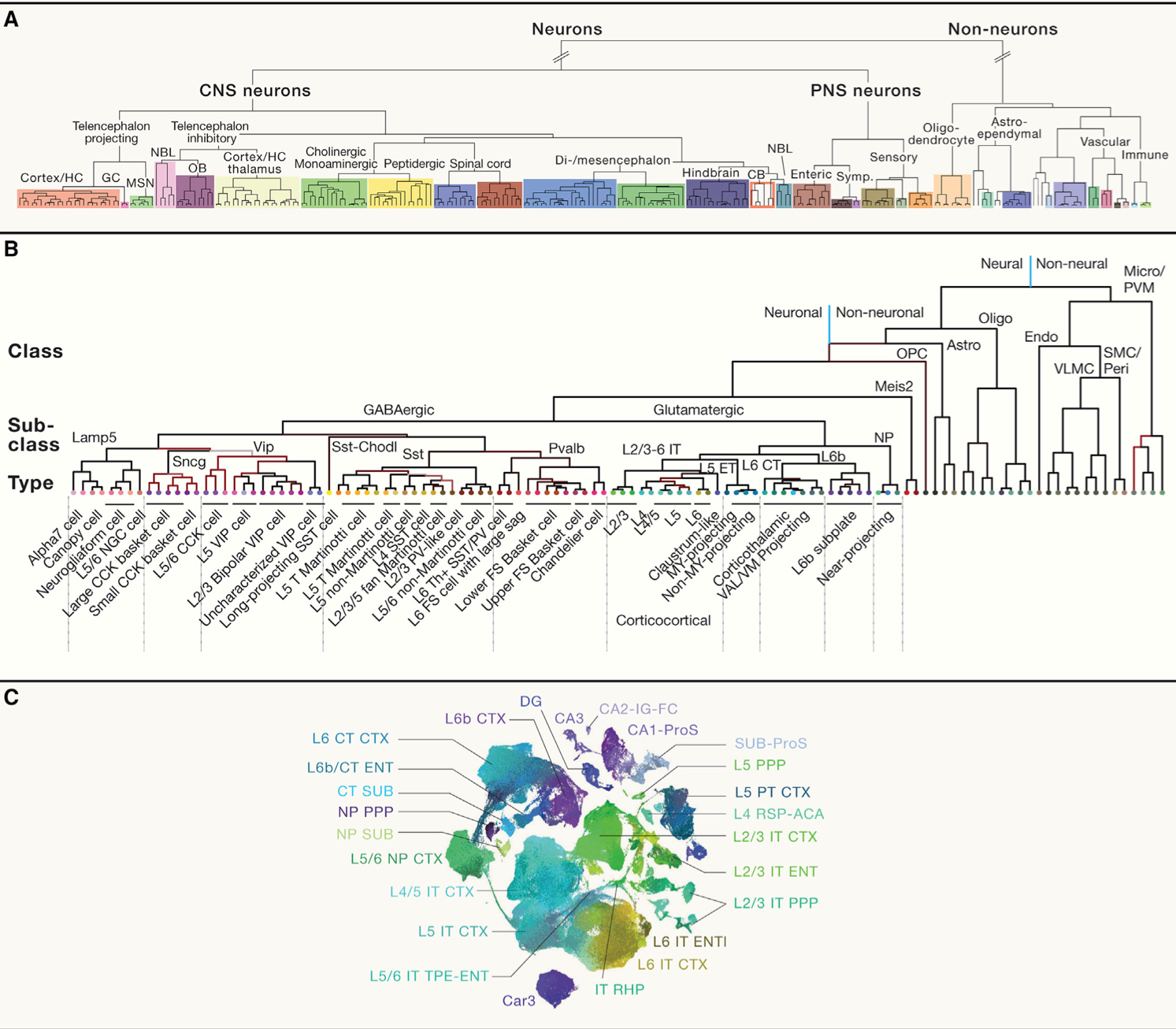
# Correspondence between transcriptomic cell types and other cellular properties
识别细胞的特性对于分别细胞类型也是很重要的,通过结合 transcriptomic 和多种其它方法来研究细胞的特征可以得出更多的结论。
其实这个小节讲的就是结合多种领域研究细胞的方法,举了很多的例子,得出了很多不同的结论。
拿他的总结来说的话就是:A transcriptomic cell type taxonomy must be linked to anotomical and functional information to evaluate the validity of the transcriptomic taxonomy and determine the appropriate level of granularity.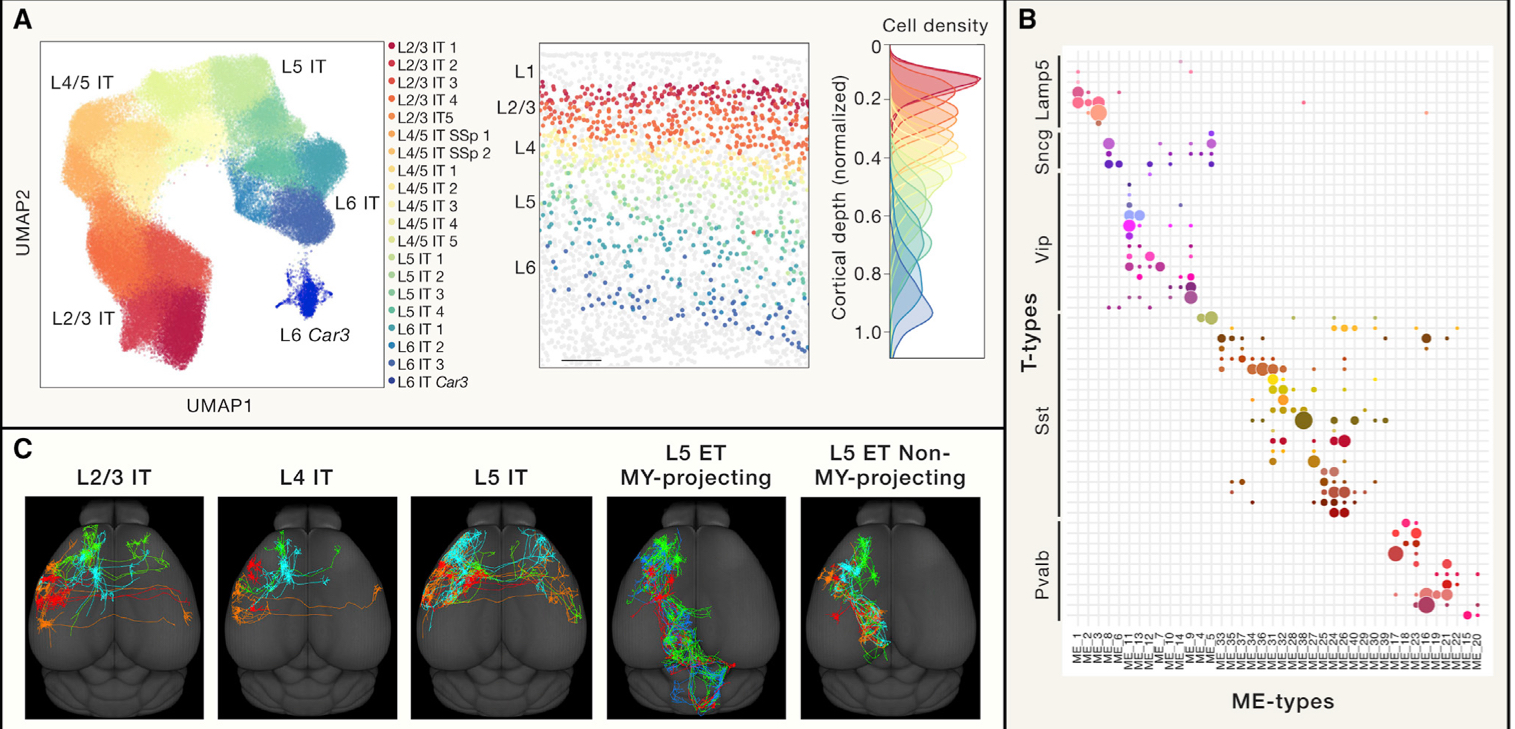
# Cell types versus cell states
细胞的类型会随着细胞的不同阶段的变化而变化
特别的,非神经细胞也会由于不同阶段分成许多不同的类,例如,Astrocytes, Oligodendrocytes, Microglia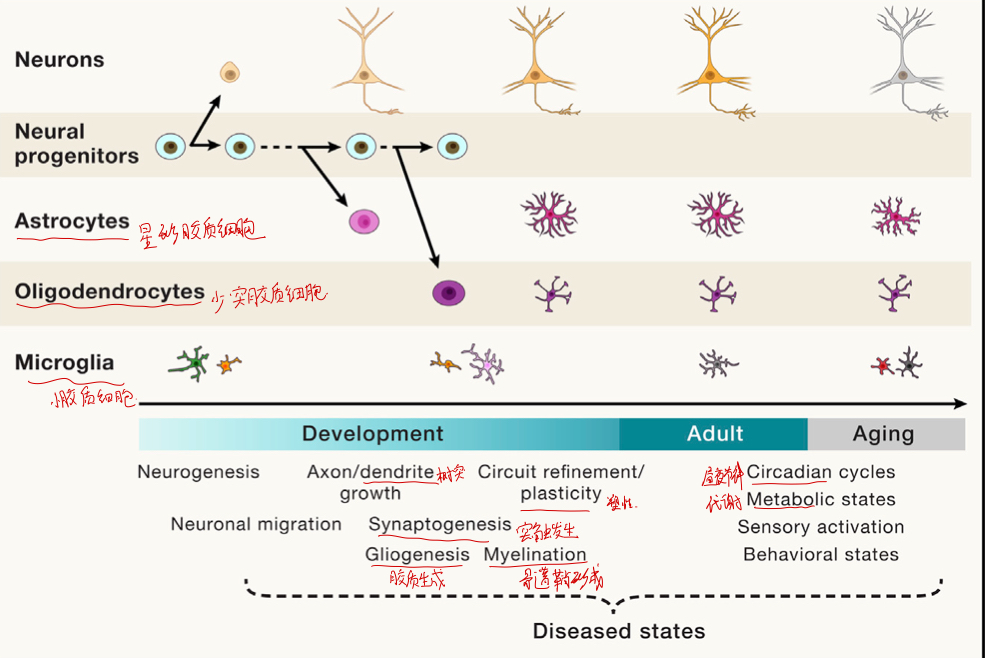
# Cell type Development
描述细胞成长过程的重要性。通过时空过程、细胞之间的交流,细胞迁移,生态的形成,最终实现功能的具体化。
然后具体拿大脑做了例子。利用各种方法可以帮助我们更好的理解成熟阶段的细胞的类型。
又具体拿 mouse cortex and spinal cord cell types 做了例子,揭示了几个常见的准则:
- First, opposing morphogen gradients establish the basic plan for cortex (anterior-posterior) or spinal cord (dorsal-ventral) patterning and provide instructive signals for the expression of complementary sets of transcription factor activators and re- pressors, which in turn define distinct neural progenitor domains within cortex or spinal cord.
- Second, driven by the transcription factor network, each type of neural progenitors generates a series of neuronal cell types. The set of cell type-defining transcrip- tion factors in a progenitor or a daughter cell can change with time, such that different neuronal types emanate from the same progenitor in a precisely timed birth order. Later in development, the same neural progenitors also generate non- neuronal cell types such as astrocytes and oligodendrocytes.
- Third, specific sets of cell adhesion molecules provide guidance cues for axon path finding and synapse formation, leading to the assembly of region- and cell type-specific local and global circuit networks.
- Fourth, patterned neural activities spontaneously emerged from the circuits and/or influenced by external inputs further sculpt synaptic connections and circuit organization to refine functional specificity of the circuit networks.
除了这些准则还介绍了细胞的类型不一定遵从一个简单的路径,它可能包括多种有分歧或聚合的分化。
# How to define cell types
总结:细胞类型是进化的产物,它们是组织最基本的功能原件。
逻辑上的第一步是用单细胞 transcriptomics-based cell type taxonomy 作为最初的框架和锚去定义细胞类型,并利用其他的细胞特性去帮助提炼细胞转录类型。
第二,我们应当综合的运用解剖的、生理学的、功能性的关于细胞转录类型的方式去定义细胞。
第三我们应当系统的学习整个细胞类型的成长过程
第四,未来综合利用它们,我们需要以概念为框架,以知识为基底去组织这些知识。("tree of cell types")
# 思考
原本我认为区分细胞是区分人类中的职业,现在我才发现这玩意怕不是类似于区分动植物界的种族。有点抽象。
# 其它补充
在 spatially resolved transcriptomics(空间转录组学) 中,in situ 方法是指直接在组织切片中进行的实验技术,用于分析不同空间位置的基因表达情况。这些术语代表了不同的技术手段,分别具有以下意义:
In situ imaging:
这是通过显微成像技术直接在组织切片上观察特定基因的表达情况。常用的技术有 ** 荧光原位杂交(FISH)** 和多重荧光成像等。这种方法使用荧光标记的探针与目标 mRNA 结合,然后通过显微镜成像,显示基因在空间位置上的分布。它提供了基因表达的定性或定量信息,结合显微镜的高分辨率,能揭示组织中基因表达的空间异质性。In situ capture:
这种技术指的是在组织切片的表面上直接捕获并固定 RNA 分子,随后进行测序分析。例如,10X Genomics 的 Visium 技术采用了一种包含空间坐标的捕获探针,将切片中的 RNA 分子捕获在特定的网格上。这种方法保留了 RNA 分子的空间信息,后续的测序分析能将这些信息还原到原始的组织切片上,生成基因表达的空间图谱。In situ sequencing:
在这种方法中,RNA 分子在原位被逆转录成 cDNA,并在组织切片上直接进行测序。最常见的技术是通过 ** 原位荧光测序(ISS)** 来读取特定位置的基因序列。这样,既保留了 RNA 分子的空间位置信息,又能够读取其序列,从而实现高分辨率的空间转录组数据生成。这一方法可以在空间上精确定位基因表达,并识别不同细胞类型或亚型。
简而言之:
- In situ imaging:在组织中通过荧光或显微镜成像观察基因表达。
- In situ capture:在组织上捕获 RNA,并保留空间位置信息进行测序。
- In situ sequencing:在组织中直接测序 RNA,既获得序列信息,又保留其空间位置。